A Minimal Package
Testing automated conversion in reticulate and wrapper classes using R6Class.
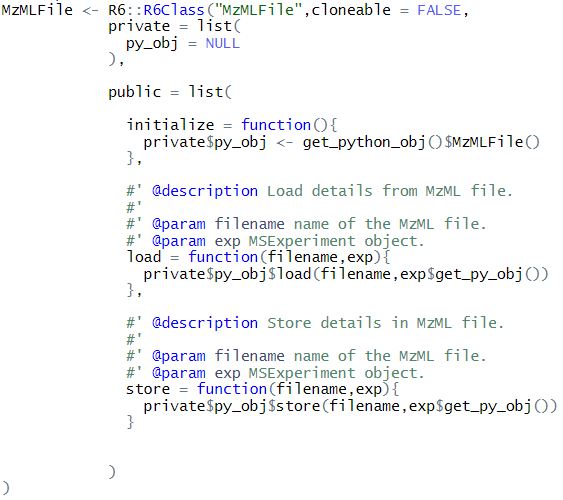
This blog features an attempt to try out the automated conversion of python to r data types and vice versa (enabled by convert = TRUE when importing pyopenms module). I have created R6 classes to wrap the corresponding pyopenms class object. The private section of this class contains the python object and the public section has wrapper functions for pyopenms class which make use of the python object. The motivation for adopting this approach was for abstraction of type conversion details and restrict the user from handling the underlying python object as much as possible.
A limited number of pyopenms classes, with selective functions, have been wrapped to demonstrate a few common use cases like file input/output, getting spectra list of an experiment and setting or extracting the peaks (m/z and Intensity values) for a spectrum.
The package is hosted here.
Note on Installation
Reticulate will automatically configure a python environment for the user when this package is loaded. If the user has no compatible version of python, they will be prompted to install Miniconda. The python dependency(pyopenms in this case) listed in Config/reticulate inside DESCRIPTION will be installed in an appropriate conda environment. The user can explicity instruct reticulate to use specific python environment having pyopenms installed by setting RETICULATE_PYTHON environment variable to a python binary, i.e. using Sys.setenv(RETICULATE_PYTHON = PATH).
Class Structure
The private section contains py_obj which has its value set at the time of object creation in initialize. get_python_obj() function returns the pyopenms module object, which is used to set py_obj as the underlying pyopenms class object. All the functions use py_obj to call the respective python function.
Currently the included classes are:
- MzMLFile,FeatureXMLFile,IdXMLFile which have load() and store() functions
- MSExperiment with functions getMSLevels(),getNrSpectra(),getSpectra(),getSpectrum(),setSpectra() and size()
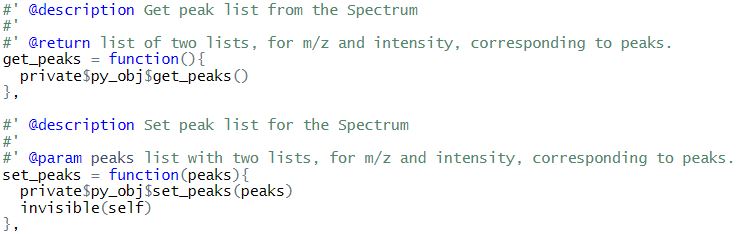
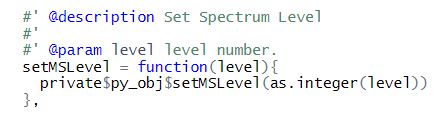
- MSSpectrum with functions getMSLevel(),setMSLevel(),getRT(),setRT(),get_peaks(),size() and set_peaks()
- FeatureMap with function getFeature() to access specific feature by index. As FeatureMap is one of the many classes which support iteration, this function was made taking the fact into account.
- Feature with getUniqueId() and getMZ()
Apart from the wrapper functions, setter and getter methods (set_py_obj() & get_py_obj()) are also present in some classes to handle the underlying python object.
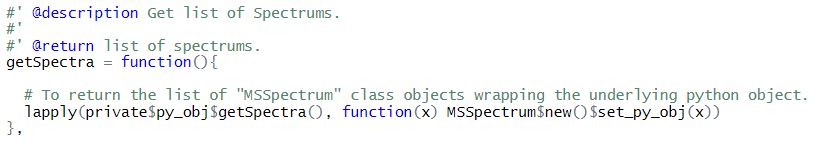
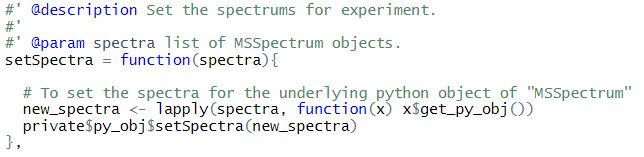
For example, consider the getSpectra() and setSpectra() functions of class MSExperiment
Using getSpectra() of python object, we get a list of MSSpectrum python objects. Then for each python object, we create an MSSpectrum object and update its underlying python class object using set_py_obj.
Here, we use get_py_obj to access the underlying python object.
One drawback of using these functions is that it will weaken the encapsulation as users can access the underlying python object.
Type Conversion.
Reticulate converts the R data types into equivalent python types when passed to a function. Similarly, when values are returned from Python to R they are converted back to R types.
We don’t need to convert the values returned from a function. But, we need to perform to and fro conversion explicitly in case where the argument passed to a function is modified.
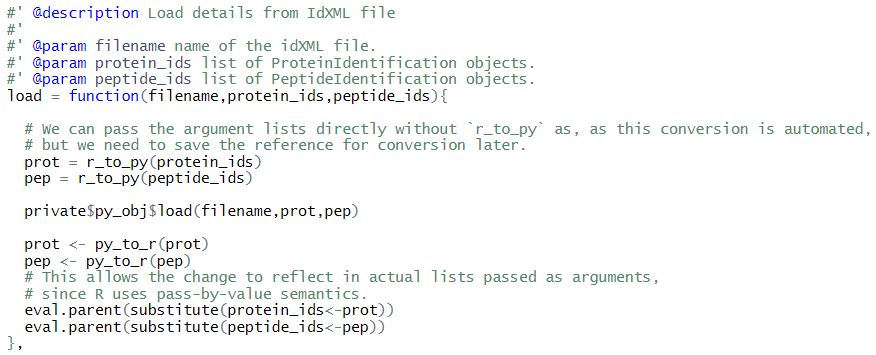
For example, consider the implementation of load() function of IdXMLFile.
Here, if protein_ids and peptide_ids lists are passed directly, then the problem is that reticulate first converts these to equivalent python lists and then passes these new objects to the function. Thus, only the converted python lists will get modified. We need to save the reference to the converted python list, in order to reflect back changes in the R list.
For setting or extracting peaks, we don’t need to do explicit conversion. The python tuple of two numpy arrays gets converted to an R list with two arrays and vice versa.
Here, using the wrapped python object we call the respective functions passing the arguments directly.
For functions using integer parameter, we need to explicitly coerce the argument to integer as otherwise reticulate will convert it to float in python since by default, the internal data type of any integer in R is double unless declared using “L” suffix.
Some code snippets used to test functionality (based on ropenms Script)
load and parse idXML file.
download.file("https://raw.githubusercontent.com/OpenMS/OpenMS/develop/share/OpenMS/examples/BSA/BSA2_OMSSA.idXML","bs.idXML")
idXML <- IdXMLFile$new()
peptids <- list()
protids <- list()
idXML$load("bs.idXML",protids,peptids)
pephits=peptids[[5]]$getHits()
pephits[[1]]$getSequence()
load and parse featureXML file.
download.file("https://raw.githubusercontent.com/OpenMS/OpenMS/develop/share/OpenMS/examples/FRACTIONS/BSA1_F1.featureXML","f.featureXML")
featXML <- FeatureXMLFile$new()
fmap <- FeatureMap$new()
featXML$load("f.featureXML",fmap)
# Accessing feature properties.
# As UniqueId is non-32 bit integer, so the conversion fails and getUniqueId() gives -1 as output.
print(paste("FeatureID:", fmap$getFeature(3)$getUniqueId()))
print(paste("FeatureID:", fmap$getFeature(3)$getMZ()))
load and parse mzML file.
download.file("https://raw.githubusercontent.com/OpenMS/OpenMS/develop/share/OpenMS/examples/BSA/BSA1.mzML","t.mzML")
mzML <- MzMLFile$new()
msexp <- MSExperiment$new()
mzML$load("t.mzML",msexp)
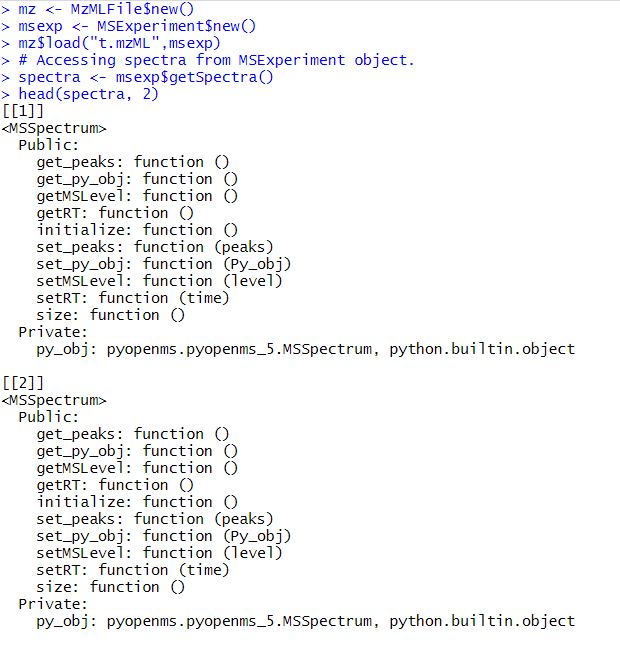
# Accessing spectra from MSExperiment object.
spectra <- msexp$getSpectra()
head(spectra, 2)
Accessing peaks for a spectrum.
spectrum <- MSSpectrum$new()
m <- seq(1000,500,-100)
intensity <- seq(50,200, length.out = length(mz))
spectrum$set_peaks(list(mz,intensity))
## Access peak with get_peaks()
do.call("cbind", spectrum$get_peaks())
Filtering ms1 spectrums and generating peak map from a spectra list.
ms1=sapply(spectra, function(x) x$getMSLevel()==1)
peaks=sapply(spectra[ms1], function(x) cbind(do.call("cbind", x$get_peaks()),x$getRT()))
peaks=do.call("rbind", peaks)
peaks_df=data.frame(peaks)
colnames(peaks_df)=c('MZ','Intensity','RT')
peaks_df$Intensity=log10(peaks_df$Intensity)
ggplot(peaks_df, aes(x=MZ, y=RT) )+geom_point(size=1, aes(colour = Intensity), alpha=0.25) + theme_minimal() + scale_colour_gradient(low = "blue", high = "yellow")
Limitations to discuss.
- The user can still access the underlying pyopenms object. So, there is not full abstraction. To see if the handling of this object should be allowed or not.
- This approach of creating wrapper R6 classes may not be very easy to automate and lead to memory intensive package.
- Although using automated conversion may eliminate explicit type conversion to a good extent, it is required when objects passed as arguments get modified.
- Many classes in pyopenms support iteration. Creating a wrapper method to iterate on the pyopenms object might be a possible solution.